plots:forest_plot_with_subgroups
Table of Contents
Forest Plot with Subgroups
Description
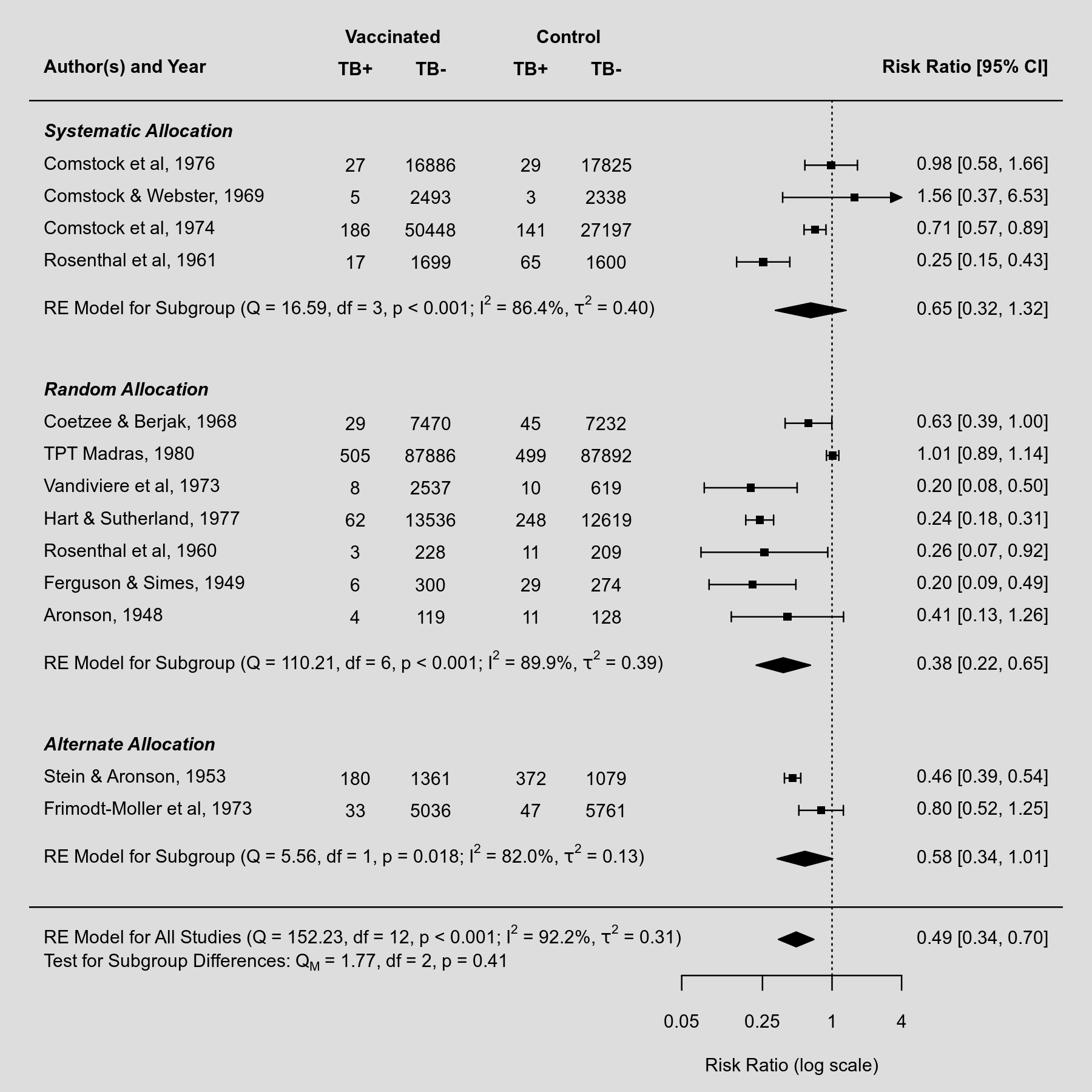
Below is an example of a forest plot with three subgroups. The results of the individual studies are shown grouped together according to their subgroup. Below each subgroup, a summary polygon shows the results when fitting a random-effects model just to the studies within that group. The summary polygon at the bottom of the plot shows the results from a random-effects model when analyzing all 13 studies.
Plot
Code
library(metafor) ### copy BCG vaccine meta-analysis data into 'dat' dat <- dat.bcg ### calculate log risk ratios and corresponding sampling variances (and use ### the 'slab' argument to store study labels as part of the data frame) dat <- escalc(measure="RR", ai=tpos, bi=tneg, ci=cpos, di=cneg, data=dat, slab=paste(author, year, sep=", ")) ### fit random-effects model res <- rma(yi, vi, data=dat) ### a little helper function to add Q-test, I^2, and tau^2 estimate info mlabfun <- function(text, x) { list(bquote(paste(.(text), " (Q = ", .(fmtx(x$QE, digits=2)), ", df = ", .(x$k - x$p), ", ", .(fmtp2(x$QEp)), "; ", I^2, " = ", .(fmtx(x$I2, digits=1)), "%, ", tau^2, " = ", .(fmtx(x$tau2, digits=2)), ")")))} ### set up forest plot (with 2x2 table counts added; the 'rows' argument is ### used to specify in which rows the outcomes will be plotted) forest(res, xlim=c(-16, 4.6), at=log(c(0.05, 0.25, 1, 4)), atransf=exp, ilab=cbind(tpos, tneg, cpos, cneg), ilab.lab=c("TB+","TB-","TB+","TB-"), ilab.xpos=c(-9.5,-8,-6,-4.5), cex=0.75, ylim=c(-2,28), top=4, order=alloc, rows=c(3:4,9:15,20:23), mlab=mlabfun("RE Model for All Studies", res), psize=1, header="Author(s) and Year") ### set font expansion factor (as in forest() above) op <- par(cex=0.75) ### add additional column headings to the plot text(c(-8.75,-5.25), 27, c("Vaccinated", "Control"), font=2) ### add text for the subgroups text(-16, c(24,16,5), pos=4, c("Systematic Allocation", "Random Allocation", "Alternate Allocation"), font=4) ### set par back to the original settings par(op) ### fit random-effects model in the three subgroups res.s <- rma(yi, vi, subset=(alloc=="systematic"), data=dat) res.r <- rma(yi, vi, subset=(alloc=="random"), data=dat) res.a <- rma(yi, vi, subset=(alloc=="alternate"), data=dat) ### add summary polygons for the three subgroups addpoly(res.s, row=18.5, mlab=mlabfun("RE Model for Subgroup", res.s)) addpoly(res.r, row= 7.5, mlab=mlabfun("RE Model for Subgroup", res.r)) addpoly(res.a, row= 1.5, mlab=mlabfun("RE Model for Subgroup", res.a)) ### fit meta-regression model to test for subgroup differences res <- rma(yi, vi, mods = ~ alloc, data=dat) ### add text for the test of subgroup differences text(-16, -1.8, pos=4, cex=0.75, bquote(paste("Test for Subgroup Differences: ", Q[M], " = ", .(fmtx(res$QM, digits=2)), ", df = ", .(res$p - 1), ", ", .(fmtp2(res$QMp)))))
plots/forest_plot_with_subgroups.txt · Last modified: by Wolfgang Viechtbauer