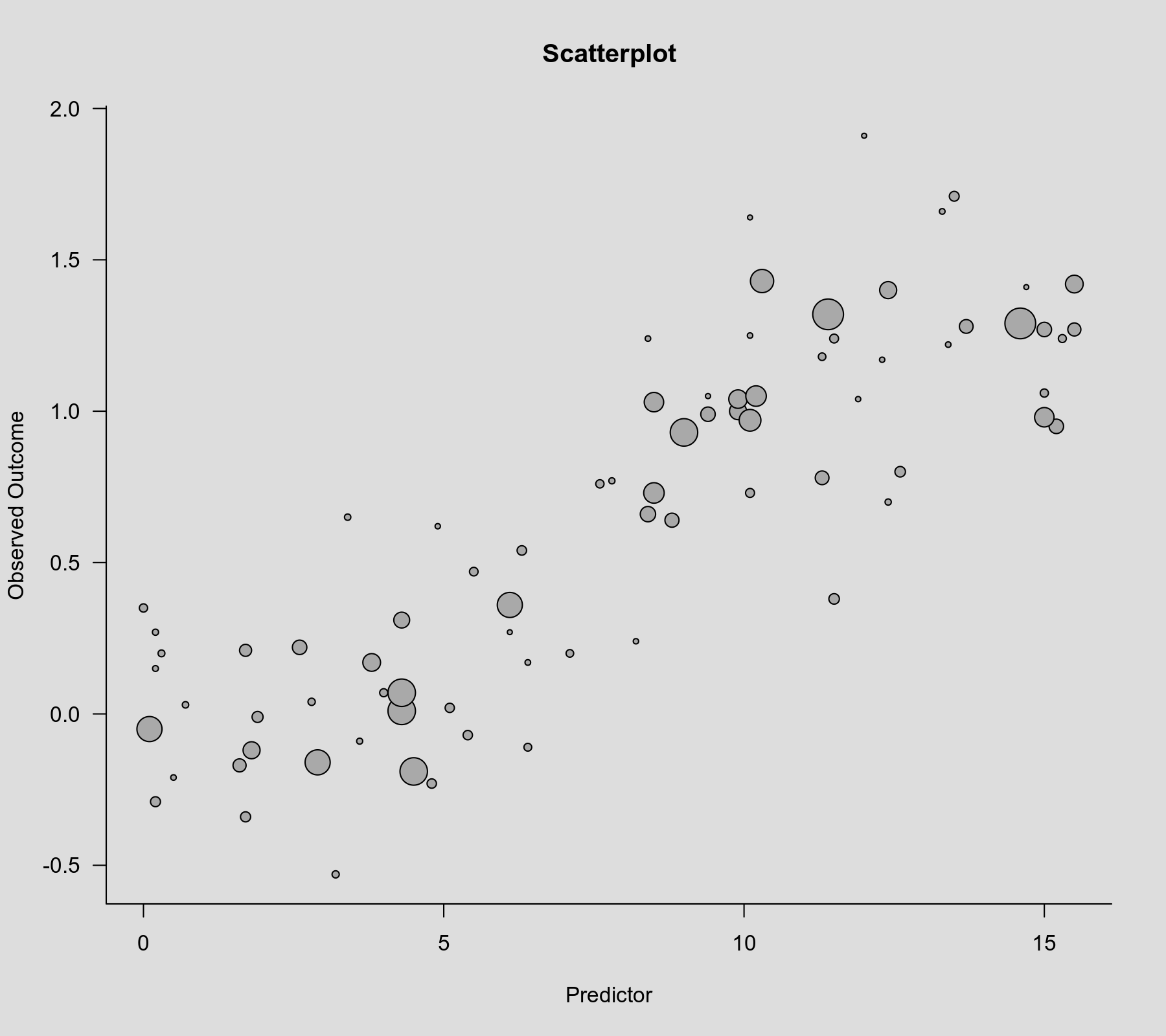
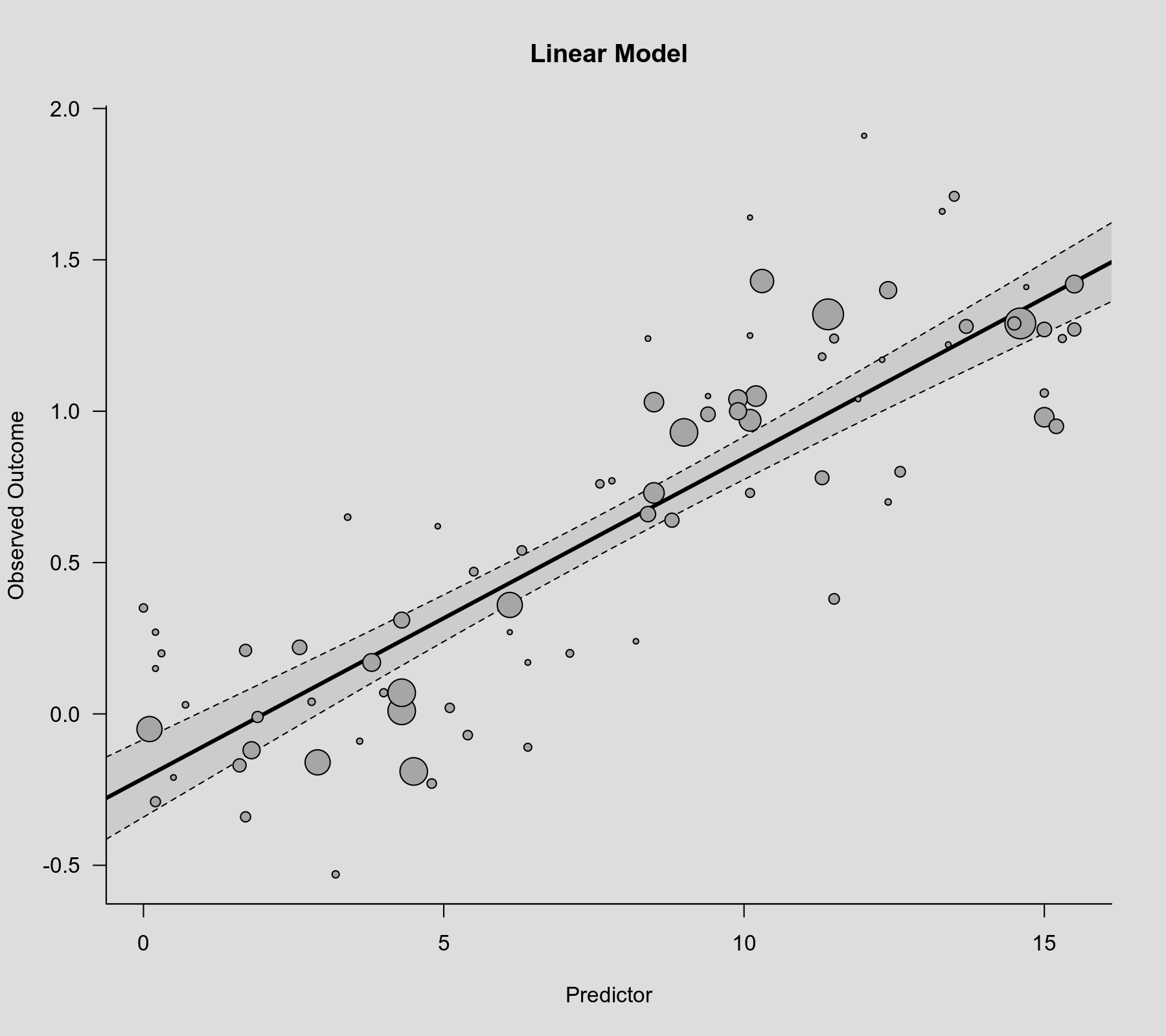
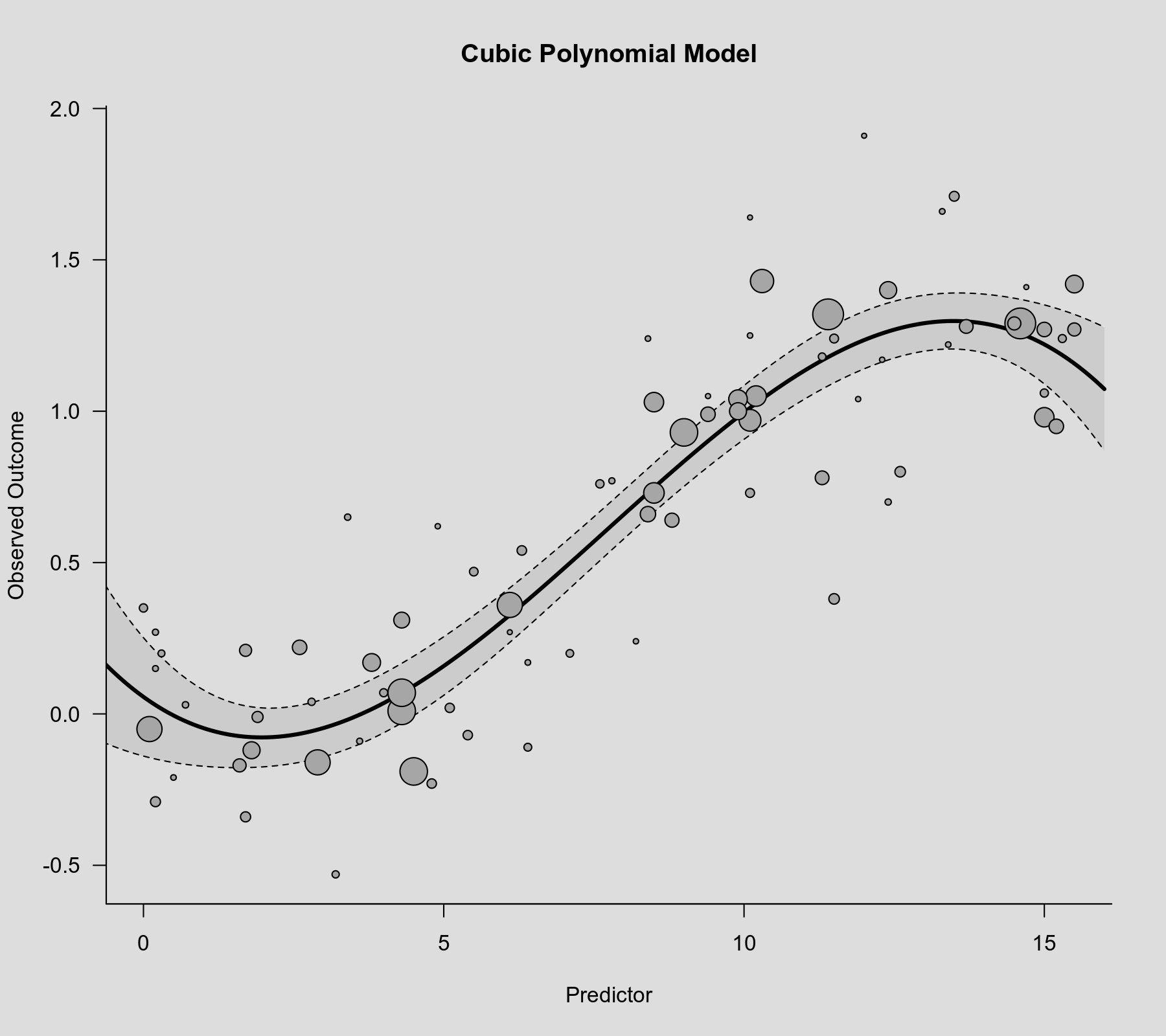
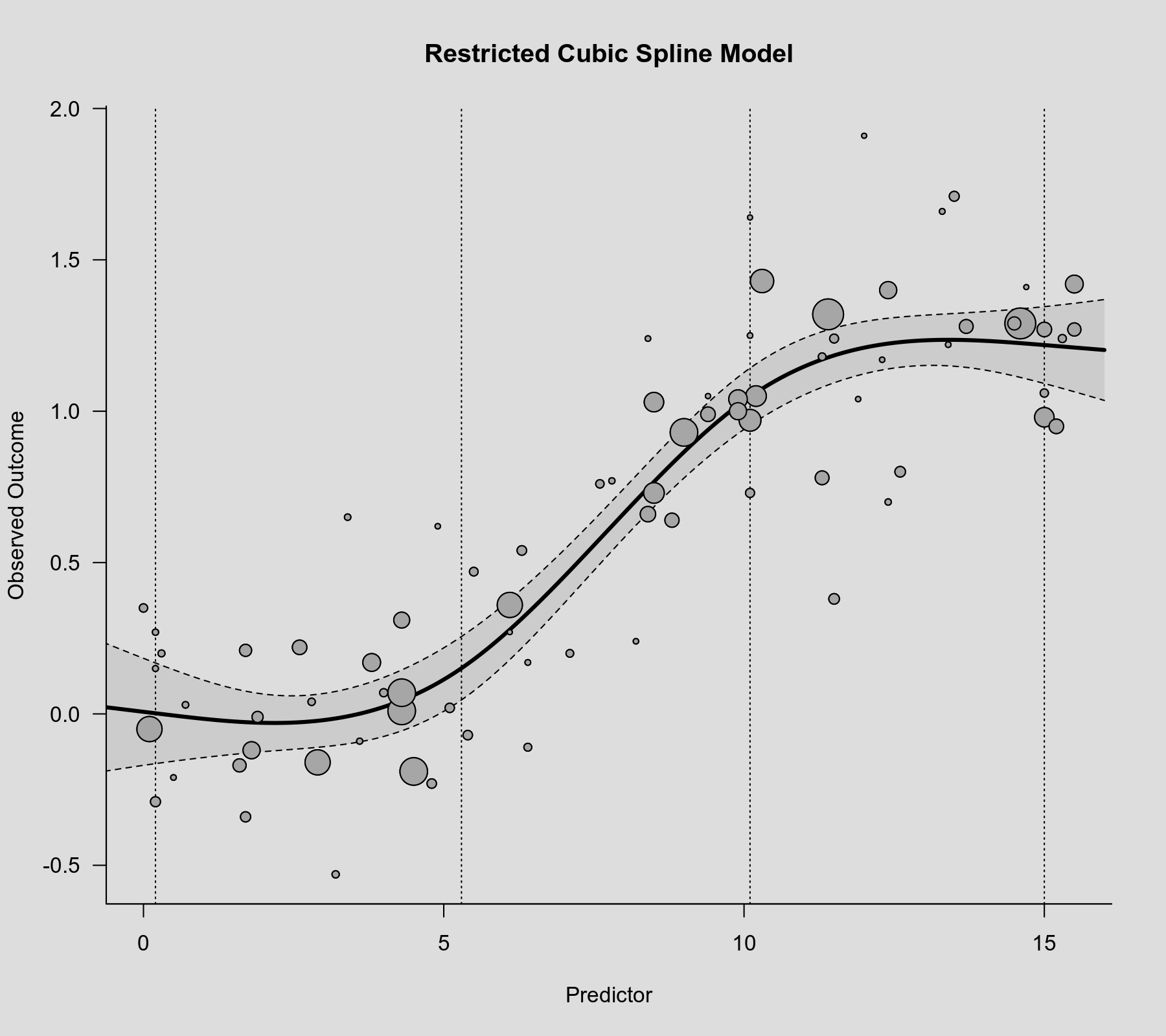
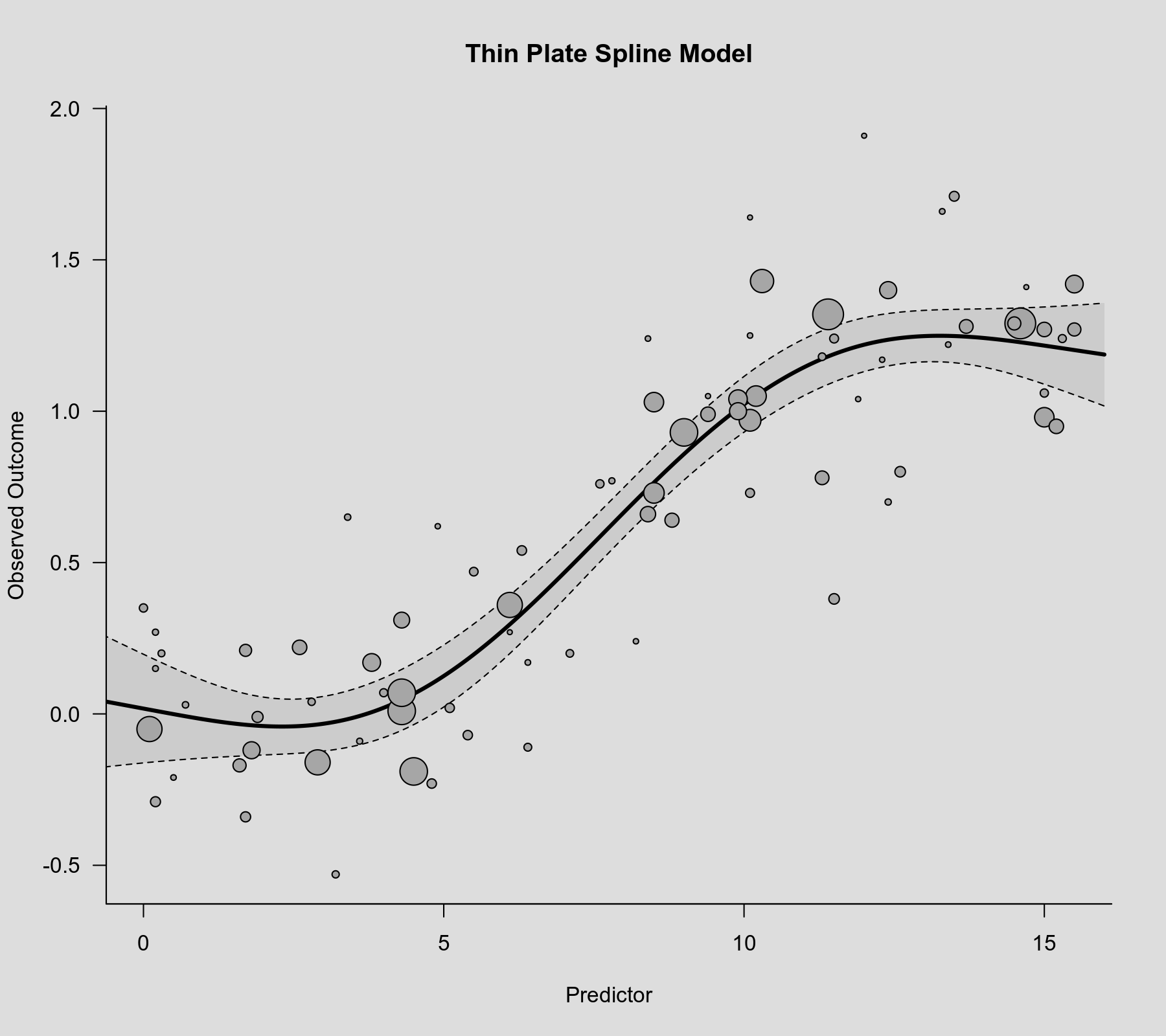
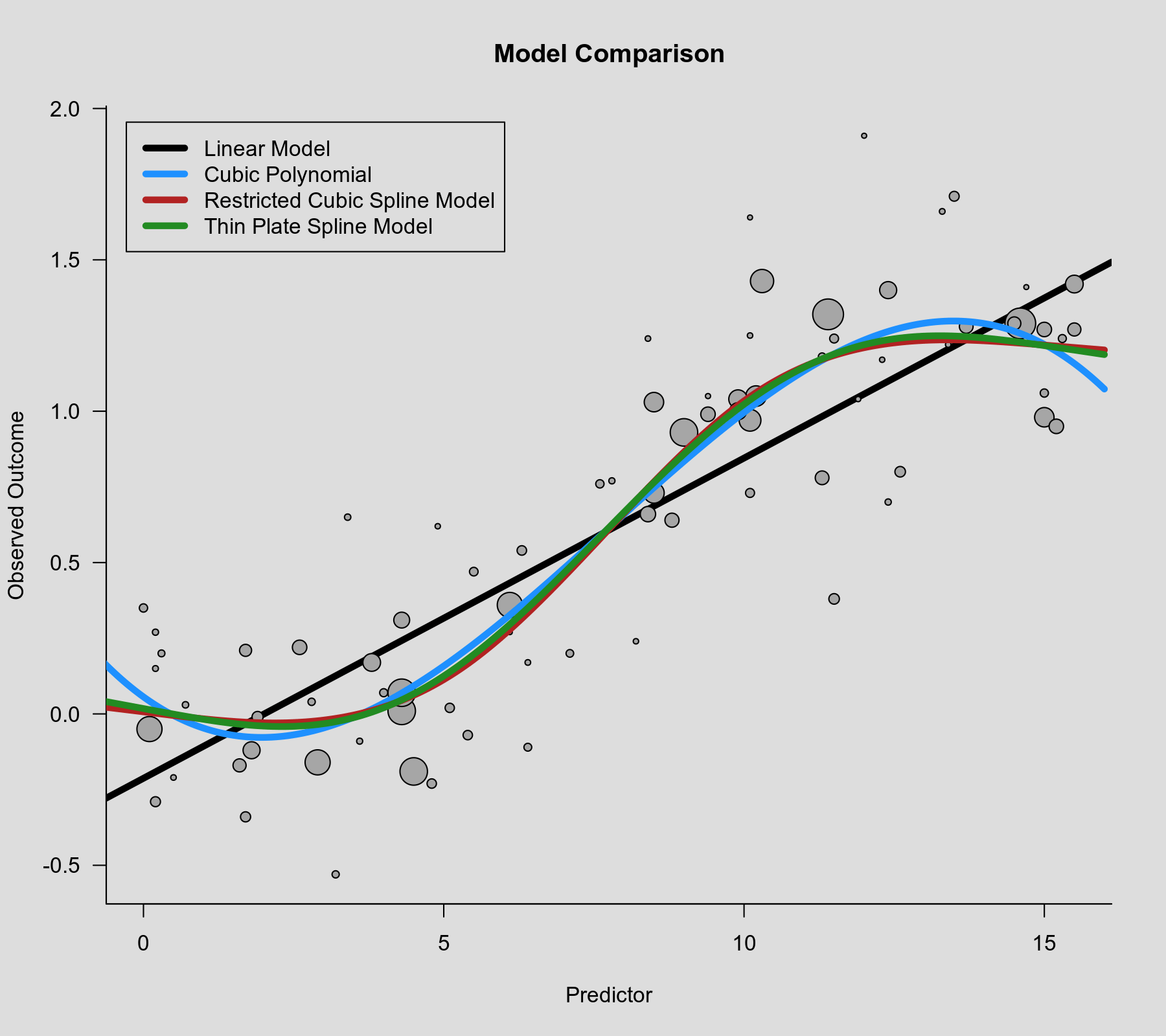
rcs() function as the second argument. Also note that rcs(xi, 4) will select the knot positions based on all (non-missing) xi values, but if there are (additional) missing values in yi and/or vi, then the actual analysis will be based on a subset of the data for which the chosen knot positions might not be ideal.