Handling Missing Data in Output/Figures
In many cases, the dataset to be used for a meta-analysis will contain studies for which insufficient information is available to compute the observed outcomes (e.g., the risk/odds ratios or the raw/standardized mean differences) or for which the values of potentially relevant moderators/covariates are unknown. We can use the dataset for the BCG vaccine meta-analysis (Colditz et al., 1994) as an illustration.
Suppose the 2×2 table data for the 6th study were not available and let's make the absolute latitude value for the 8th study missing:
library(metafor) dat.bcg[6,4:7] <- NA dat.bcg$ablat[8] <- NA dat.bcg
trial author year tpos tneg cpos cneg ablat alloc 1 1 Aronson 1948 4 119 11 128 44 random 2 2 Ferguson & Simes 1949 6 300 29 274 55 random 3 3 Rosenthal et al 1960 3 228 11 209 42 random 4 4 Hart & Sutherland 1977 62 13536 248 12619 52 random 5 5 Frimodt-Moller et al 1973 33 5036 47 5761 13 alternate 6 6 Stein & Aronson 1953 NA NA NA NA 44 alternate 7 7 Vandiviere et al 1973 8 2537 10 619 19 random 8 8 TPT Madras 1980 505 87886 499 87892 NA random 9 9 Coetzee & Berjak 1968 29 7470 45 7232 27 random 10 10 Rosenthal et al 1961 17 1699 65 1600 42 systematic 11 11 Comstock et al 1974 186 50448 141 27197 18 systematic 12 12 Comstock & Webster 1969 5 2493 3 2338 33 systematic 13 13 Comstock et al 1976 27 16886 29 17825 33 systematic
Now suppose we want to fit a random-effects model to these data, using the (log) risk ratio as the outcome measure. We can do this with:
dat <- escalc(measure="RR", ai=tpos, bi=tneg, ci=cpos, di=cneg, data=dat.bcg) res <- rma(yi, vi, data=dat, slab=paste0(trial, ") ", author, ", ", year))
A warning will be issued by the rma function, since the log risk ratio could not be computed for the 6th study, leading to missing data (NA values):
Warning message: In rma(measure = "RR", ai = tpos, bi = tneg, ci = cpos, di = cneg, : Studies with NAs omitted from model fitting.
We can now draw a forest plot based on the results from the meta-analysis with:
forest(res, atransf=exp, at=log(c(0.05, 0.25, 1, 4)), xlim=c(-9,6))
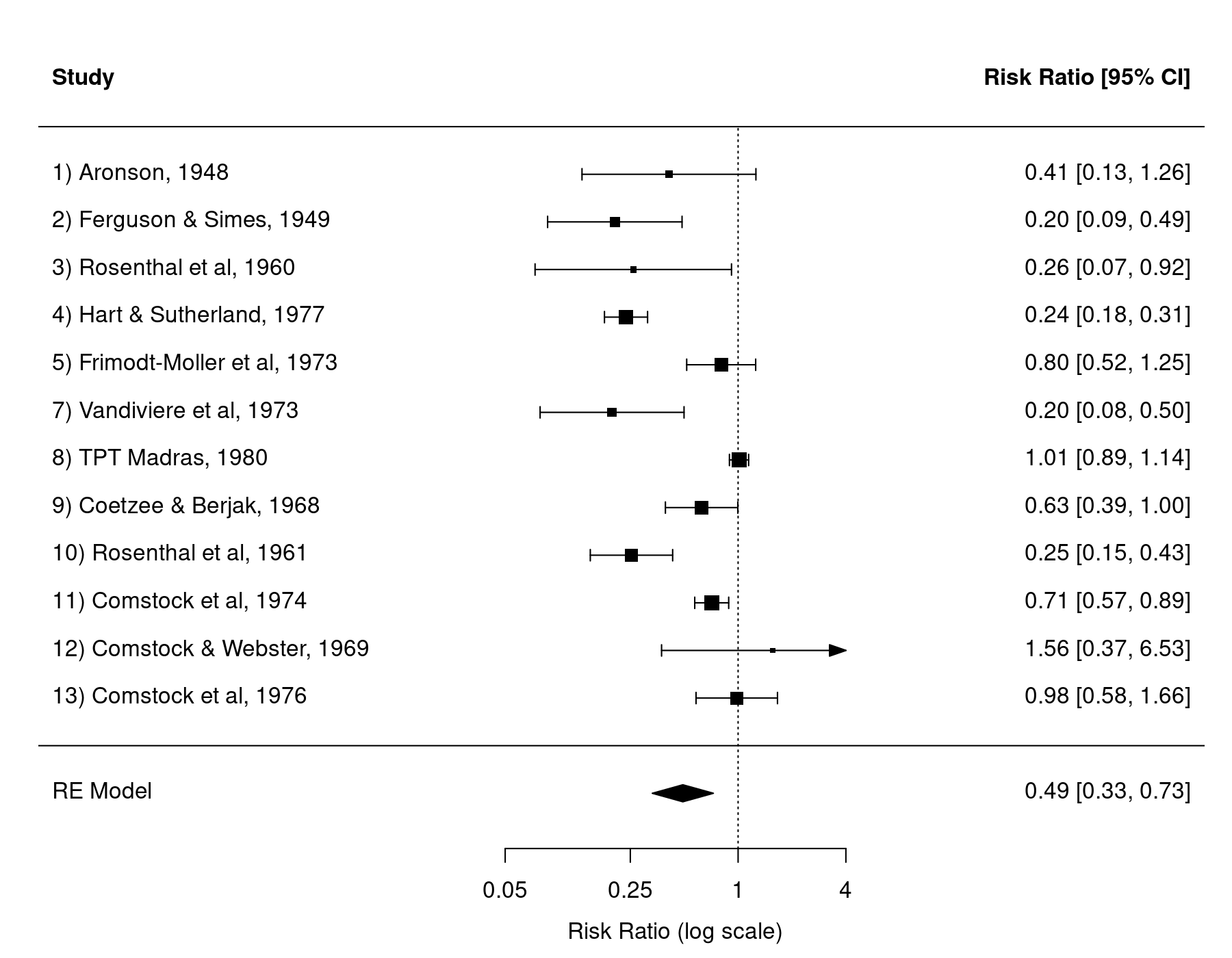
As we can see, the 6th study has been omitted from the forest plot. However, we may still want to have the study included in the forest plot (e.g., as a visual cue that the study did in fact exist). This is possible by changing the na.action option under the global options (see help(options) for more details on options settings). In particular, the default setting in R for na.action is na.omit, which results in the behavior shown above.
We can change this with:
options(na.action = "na.pass")
Various functions in the metafor package will now let missing values pass on to the output and figures. We can now redraw the forest plot:
forest(res, atransf=exp, at=log(c(0.05, 0.25, 1, 4)), xlim=c(-9,6))

And now, the 6th study is shown in the plot (of course, since the log risk ratio and corresponding CI cannot be computed, the actual results remain missing).
When inspecting output, this issue may also be relevant. For example, suppose we fit a mixed-effects meta-regression model to the log risk ratios using the absolute latitude of the study location as a potential moderator:
res <- rma(yi, vi, mods = ~ ablat, data=dat)
Again, a warning will be issued, since parts of the data were missing (i.e., the yi and vi values for the 6th study and the ablat value for the 8th study).
Now we want to inspect the studentized residuals for these studies. The default behavior (with the na.action set to na.omit) would be:
options(na.action = "na.omit") rstudent(res)
resid se z 1 0.2230 0.7158 0.3115 2 -0.2439 0.6860 -0.3555 3 -0.3290 0.7692 -0.4277 4 -0.2053 0.5610 -0.3660 5 0.0198 0.6019 0.0329 7 -1.3702 0.5473 -2.5036 9 0.1728 0.5182 0.3334 10 -0.3928 0.5069 -0.7750 11 0.0442 0.5377 0.0822 12 1.2682 0.8147 1.5566 13 0.8292 0.2767 2.9962
Note that studies 6 and 8 are not shown in the output.
If we want to have vectors of the same length as the original data, we could use:
options(na.action = "na.pass") rstudent(res)
resid se z 1 0.2230 0.7158 0.3115 2 -0.2439 0.6860 -0.3555 3 -0.3290 0.7692 -0.4277 4 -0.2053 0.5610 -0.3660 5 0.0198 0.6019 0.0329 6 NA NA NA 7 -1.3702 0.5473 -2.5036 8 NA NA NA 9 0.1728 0.5182 0.3334 10 -0.3928 0.5069 -0.7750 11 0.0442 0.5377 0.0822 12 1.2682 0.8147 1.5566 13 0.8292 0.2767 2.9962
References
Colditz, G. A., Brewer, T. F., Berkey, C. S., Wilson, M. E., Burdick, E., Fineberg, H. V., et al. (1994). Efficacy of BCG vaccine in the prevention of tuberculosis: Meta-analysis of the published literature. Journal of the American Medical Association, 271(9), 698–702.